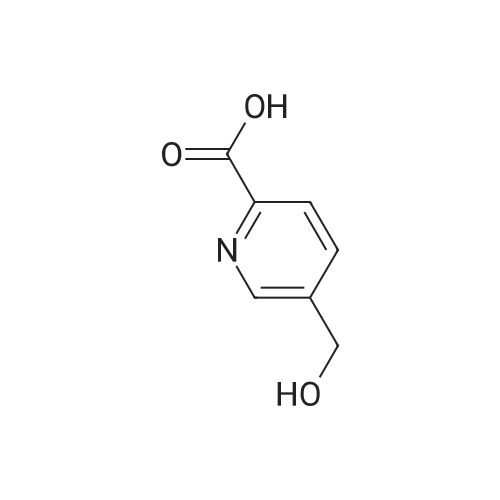
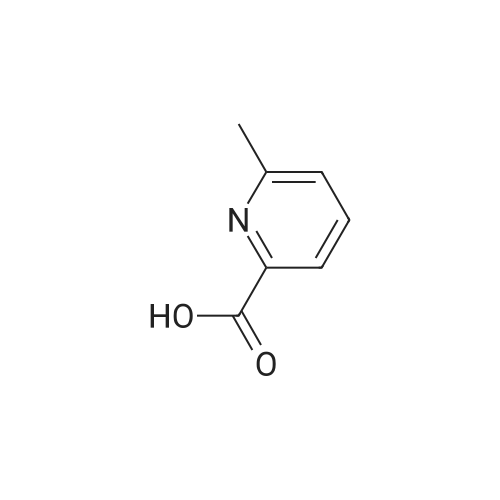
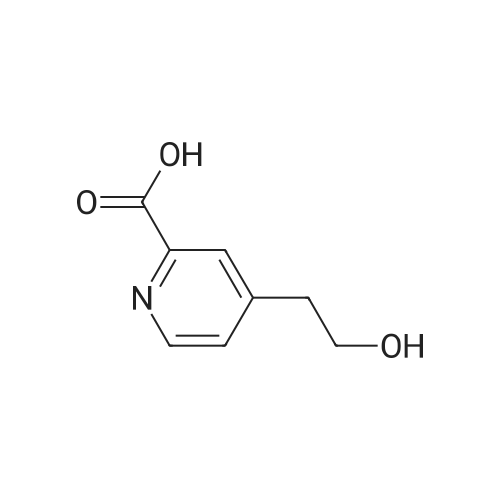
Synthesis of bitopic ligands based on fallypride and evaluation of their affinity and selectivity towards dopamine D2 and D3 receptors
Tian, Gui-Long
;
Hsieh, Chia-Ju
;
Taylor, Michelle
, et al.
Eur. J. Med. Chem.,2023,261,115751.
DOI:
10.1016/j.ejmech.2023.115751
PubMed ID:
37688938
More
Abstract: The difference in the secondary binding site (SBS) between the dopamine 2 receptor (D2R) and dopamine 3 receptor (D3R) has been used in the design of compounds displaying selectivity for the D3R versus D2R. In the current study, a series of bitopic ligands based on Fallypride were prepared with various secondary binding fragments (SBFs) as a means of improving the selectivity of this benzamide analog for D3R versus D2R. We observed that compounds having a small alkyl group with a heteroatom led to an improvement in D3R versus D2R selectivity. Increasing the steric bulk in the SBF increase the distance between the pyrrolidine N and Asp110, thereby reducing D3R affinity. The best-in-series compound was (2S,4R)-trans-27 which had a modest selectivity for D3R versus D2R and a high potency in the β-arrestin competition assay which provides a measure of the ability of the compound to compete with endogenous dopamine for binding to the D3R. The results of this study identified factors one should consider when designing bitopic ligands based on Fallypride displaying an improved affinity for D3R versus D2R.
Keywords:
Dopamine 2 receptor ;
Dopamine 3 receptor ;
Fallypride ;
Bitopic ligands ;
PET imaging
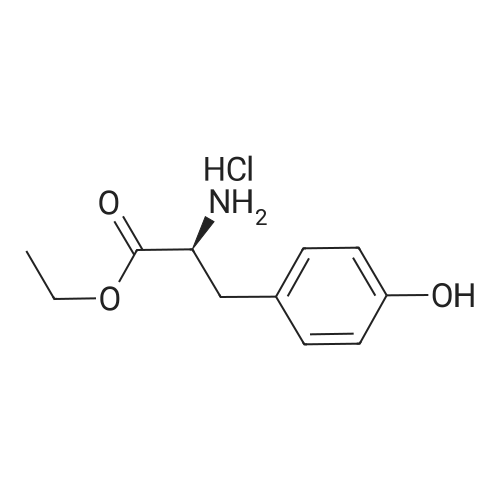
Purchased from AmBeed:
89031-84-5 ;
40216-83-9 ;
88-13-1 ;
135042-17-0 ;
110-52-1 ;
488-93-7 ;
18742-02-4 ;
98-98-6 ;
86864-60-0 ;
62-23-7 ;
29886-64-4 ;
59741-04-7 ;
4385-62-0 ;
15268-31-2 ;
134441-72-8 ;
54751-01-8 ;
166173-78-0 ;
213131-32-9 ;
98-98-6 ;
69966-55-8 ;
154342-67-3
...More
Structure activity relationship of pyrazinoic acid analogs as potential antimycobacterial agents
Hegde, Pooja V.
;
Aragaw, Wassihun W.
;
Cole, Malcolm S.
, et al.
Bioorgan. Med. Chem.,2022,74,117046.
DOI:
10.1016/j.bmc.2022.117046
PubMed ID:
36228522
More
Abstract: Tuberculosis (TB) remains a leading cause of infectious disease-related mortality and morbidity. Pyrazinamide (PZA) is a critical component of the first-line TB treatment regimen because of its sterilizing activity against non-replicating Mycobacterium tuberculosis (Mtb), but its mechanism of action has remained enigmatic. PZA is a prodrug converted by pyrazinamidase encoded by pncA within Mtb to the active moiety, pyrazinoic acid (POA) and PZA resistance is caused by loss-of-function mutations to pyrazinamidase. We have recently shown that POA induces targeted protein degradation of the enzyme PanD, a crucial component of the CoA biosynthetic pathway essential in Mtb. Based on the newly identified mechanism of action of POA, along with the crystal structure of PanD bound to POA, we designed several POA analogs using structure for interpretation to improve potency and overcome PZA resistance. We prepared and tested ring and carboxylic acid bioisosteres as well as 3, 5, 6 substitutions on the ring to study the structure activity relationships of the POA scaffold. All the analogs were evaluated for their whole cell antimycobacterial activity, and a few representative mols. were evaluated for their binding affinity, towards PanD, through isothermal titration calorimetry. We report that analogs with ring and carboxylic acid bioisosteres did not significantly enhance the antimicrobial activity, whereas the alkylamino-group substitutions at the 3 and 5 position of POA were found to be up to 5 to 10-fold more potent than POA. Further development and mechanistic anal. of these analogs may lead to a next generation POA analog for treating TB.
Keywords:
Tuberculosis ;
Pyrazinoic acid ;
pyrazinamide
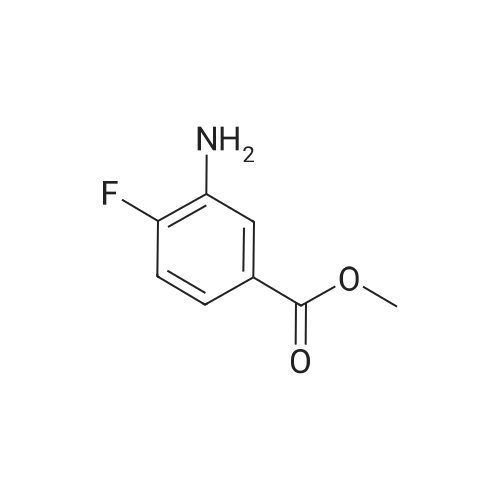
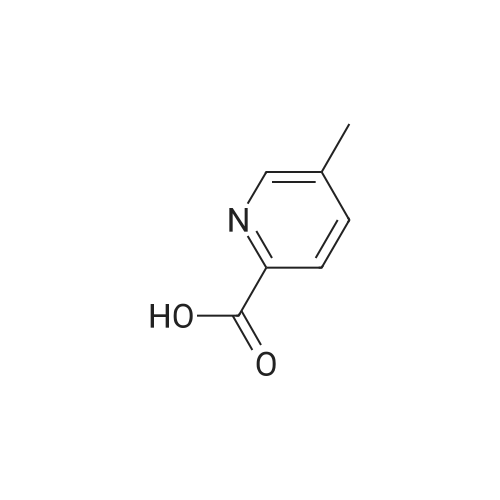
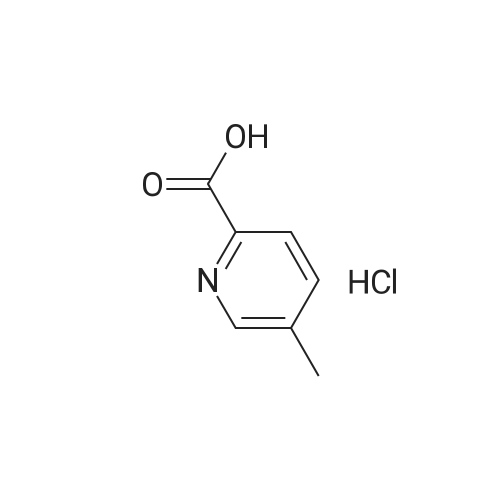
Purchased from AmBeed:
16289-54-6 ;
5521-55-1 ;
22047-25-2 ;
98-80-6 ;
40155-47-3 ;
5720-05-8 ;
879-65-2 ;
98-96-4 ;
31519-62-7 ;
23688-89-3 ;
23611-75-8 ;
33332-25-1 ;
20737-42-2 ;
61442-38-4 ;
17933-03-8 ;
50681-25-9 ;
13924-99-7 ;
40155-43-9 ;
36070-80-1 ;
4595-61-3 ;
118853-60-4 ;
41110-28-5 ;
40155-42-8 ;
937669-80-2 ;
98-98-6 ;
31462-59-6 ;
16419-60-6 ;
5424-01-1 ;
59-67-6 ;
34604-60-9 ;
27398-39-6 ;
1196151-53-7 ;
19847-12-2 ;
13965-03-2 ;
876161-05-6 ;
27825-21-4 ;
2164-61-6 ;
4604-72-2 ;
98-97-5 ;
24005-61-6 ;
103-67-3 ;
5521-61-9 ;
2516-34-9 ;
2719-27-9 ;
123-90-0 ;
6761-50-8 ;
625-43-4 ;
872-64-0 ;
36932-49-7 ;
1528085-68-8 ;
1195533-51-7 ;
13534-79-7
...More

 Chemistry
Chemistry
 Pharmaceutical Intermediates
Pharmaceutical Intermediates
 Inhibitors/Agonists
Inhibitors/Agonists
 Material Science
Material Science














 For Research Only
For Research Only
 120K+ Compounds
120K+ Compounds
 Competitive Price
Competitive Price
 1-2 Day Shipping
1-2 Day Shipping