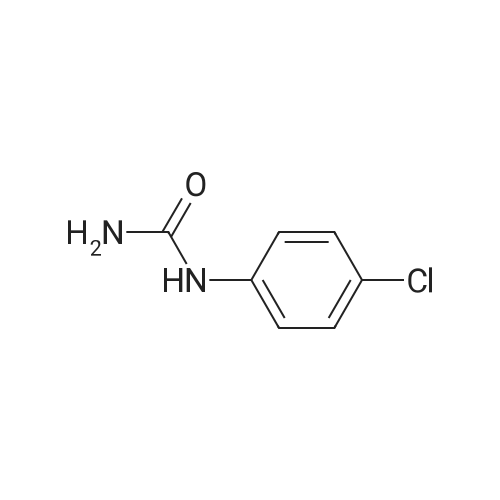
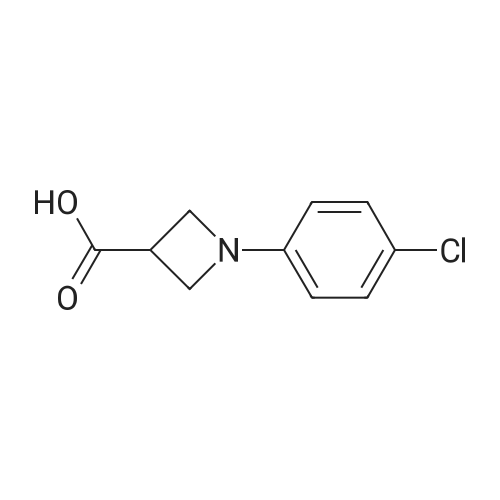
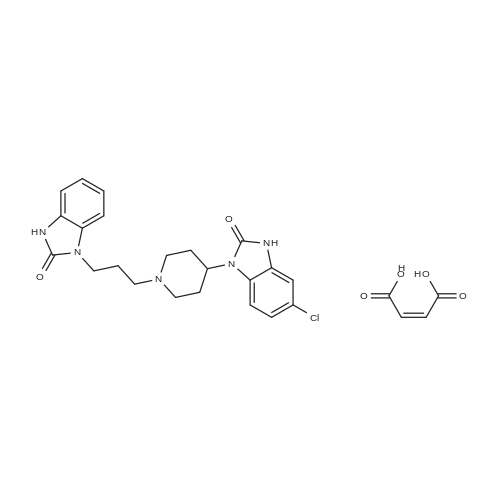
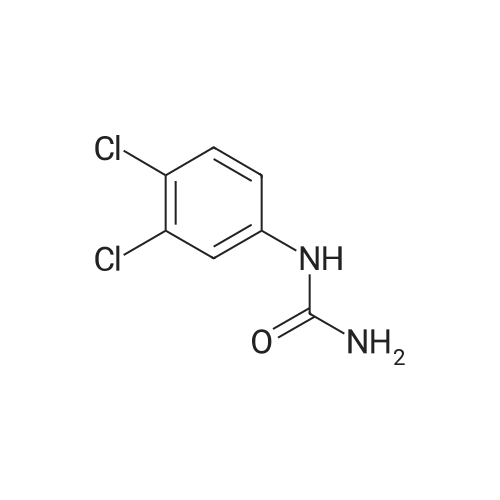
|
With potassium hydroxide; In water; at 50℃;pH 11.0;Product distribution / selectivity; |
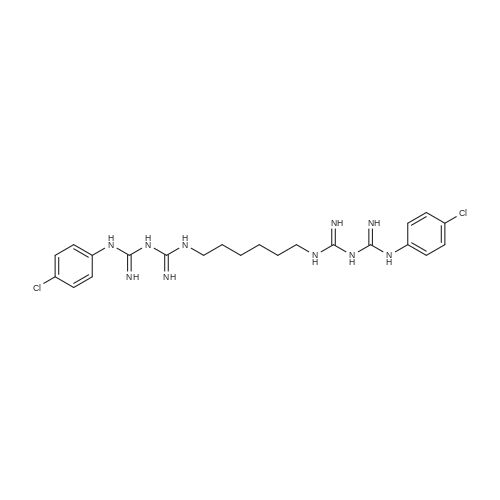
Chlorhexidine (C22H30N10Cl2), obtained commercially, was reacted with sodium hydroxide to form chlorhexidine dihydrate (C22H30N10Cl2.1.3H2O). Approximately 100 g of a starting material <strong>[56-95-1]chlorhexidine diacetate</strong> was dissolved in 1300 mL of warm deionized water at approximately 50 C. 6 M potassium hydroxide (KOH) was added drop-wise with stirring. A precipitate formed immediately and continued to form upon addition of base until the solution reached a pH of 11. The precipitate was filtered and washed six times with warm, 50 C., deionized water, and then dried in an oven at 60 C. to produce approximately 78 g of chlorhexidine dihydrate. These compounds were analyzed using energy dispersive x-ray spectroscopy (EDX), Fourier transform infrared spectroscopy (FTIR), thermogravimetric analysis (TGA), and proton nuclear magnetic resonance (1H NMR),EDXChlorhexidine and chlorhexidine dihydrate were analyzed using EDX, a technique well known to those of skill in the art. Table 1 provides both the theoretical and actual elemental composition of chlorhexidine and chlorhexidine dihydrate obtained from the EDX analysis.; FTIRFTIR was used to compare the characteristic peaks of different functional groups in chlorhexidine dihydrate and chlorhexidine. Chlorhexidine had peaks at 3513, 3473, 3410, 3371 cm-1, characteristic of N-H stretching, and peaks at 1635 and 1595 cm-1, characteristic of aromatic and aliphatic guanidine absorptions (ArNHC(N-H)NHAr) and ((CH3)2NC(N-H)C(CH3)2). The chlorhexidine dihydrate spectrum of FIG. 3 had peaks at 3458 and 3406 cm-1, characteristic of N-H stretching. The decreased frequencies likely were attributable to hydrogen bonding. The chlorhexidine dihydrate spectrum also had a broad band between 3300-2850 cm-1 that was characteristic of an intermolecular OH hydrogen-bonded bridge (typically appearing between 3405 and 2936 cm-1). Chlorhexidine dihydrate also had the aromatic guanadine peak at 1605 cm-1. The decreased frequency, again, likely was attributable to hydrogen bonding.TGATGA was used to determine the moisture content of chlorhexidine base (FIG. 4) and chlorhexidine dehydrate (FIG. 5). As shown by the derivative weight loss curve of FIG. 5, there was a loss of a small molecule (presumably water) at 100 C. and a mass decrease of 4.700% at 120.07 C. for chlorhexidine dihydrate. The mass loss likely corresponded to the 3.98% water present in the chlorhexidine dihydrate.1H NMRProton nuclear magnetic resonance (1H NMR) spectroscopy was used to analyze the structure of chlorhexidine dihydrate. The 1H NMR spectrum of chlorhexidine (FIG. 6) had peaks at 8.5, 7.25, 7.0, 3.3, 3.15, 1.9, 1.6, 1.4, and 1.25 ppm. The 1H NMR spectrum of chlorhexidine dihydrate (FIG. 7) had peaks at 8.5, 7.2, 6.9, 3.3, 3.15, 1.85, 1.6, 1.35, and 1.25 ppm, similar to that of chlorhexidine. The intensities, however, were different. Specifically, the peak at 8.5 ppm was significantly less intense in the chlorhexidine dihydrate spectrum. The peaks at 8.5, 1.85, and 1.35 ppm showed no spin-spin coupling and were therefore in rapid equilibrium in the deuterated methanol solvent (tautomerization). The water appeared to preferentially stabilize some of the tautomers of chlorhexidine. |

 Chemistry
Chemistry
 Pharmaceutical Intermediates
Pharmaceutical Intermediates
 Inhibitors/Agonists
Inhibitors/Agonists
 Material Science
Material Science














 For Research Only
For Research Only
 120K+ Compounds
120K+ Compounds
 Competitive Price
Competitive Price
 1-2 Day Shipping
1-2 Day Shipping