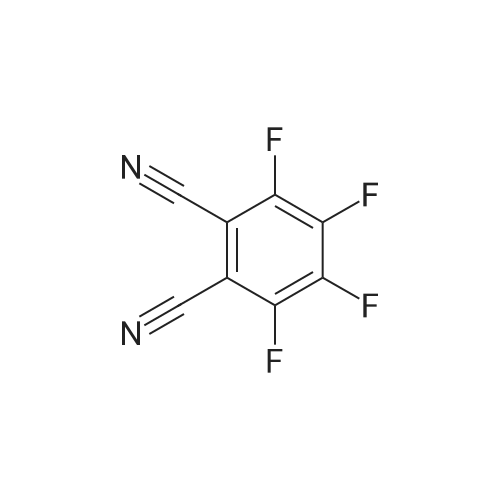
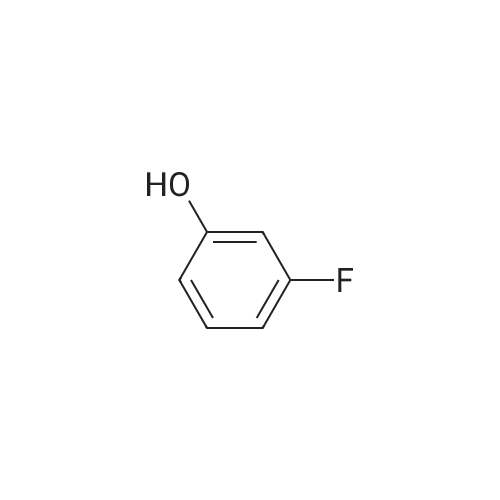
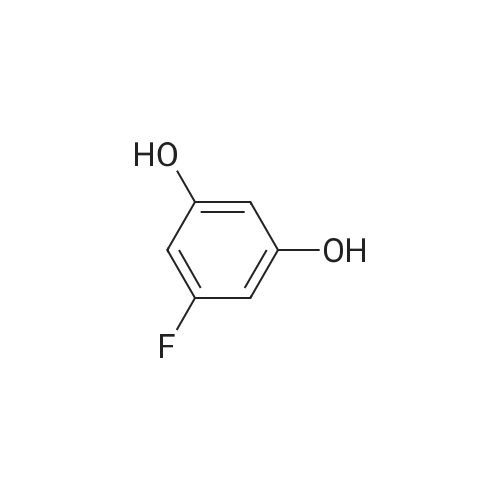
Azapeptides with unique covalent warheads as SARS-CoV-2 main protease inhibitors
Kaustav Khatua
;
Yugendar R. Alugubelli
;
Kai S. Yang
, et al.
Antivir. Res.,2024,225,105874.
DOI:
10.1016/j.antiviral.2024.105874
More
Abstract: The main protease (MPro) of SARS-CoV-2, the causative agent of COVID-19, is a pivotal nonstructural protein critical for viral replication and pathogenesis. Its protease function relies on three active site pockets for substrate recognition and a catalytic cysteine for enzymatic activity. To develop potential SARS-CoV-2 antivirals, we successfully synthesized a diverse range of azapeptide inhibitors with various covalent warheads to target MPro's catalytic cysteine. Our characterization identified potent MPro inhibitors, including MPI89 that features an aza-2,2-dichloroacetyl warhead with a remarkable EC50 value of 10 nM against SARS-CoV-2 infection in ACE2+ A549 cells and a selective index of 875. MPI89 is also remarkably selective and shows no potency against SARS-CoV-2 papain-like protease and several human proteases. Crystallography analyses demonstrated that these inhibitors covalently engaged the catalytic cysteine and used the aza-amide carbonyl oxygen to bind to the oxyanion hole. MPI89 stands as one of the most potent MPro inhibitors, suggesting the potential for further exploration of azapeptides and the aza-2,2-dichloroacetyl warhead for developing effective therapeutics against COVID-19.
Keywords:
COVID-19 ;
SARS-CoV-2 ;
Main protease ;
Azapeptide ;
Covalent inhibitor
Purchased from AmBeed:
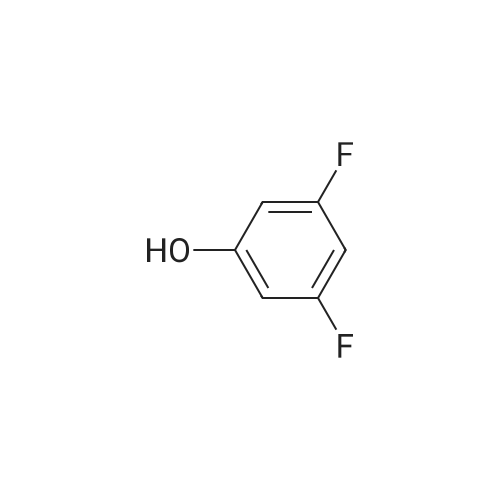
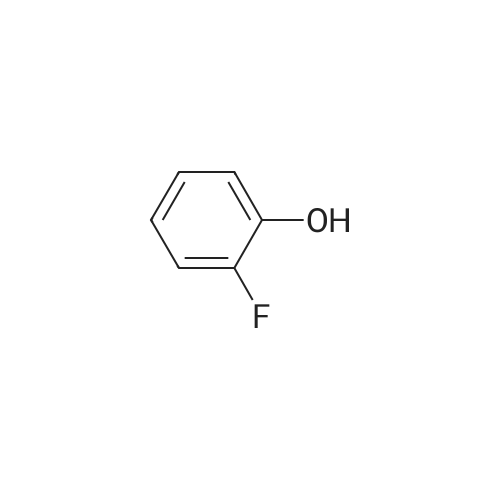
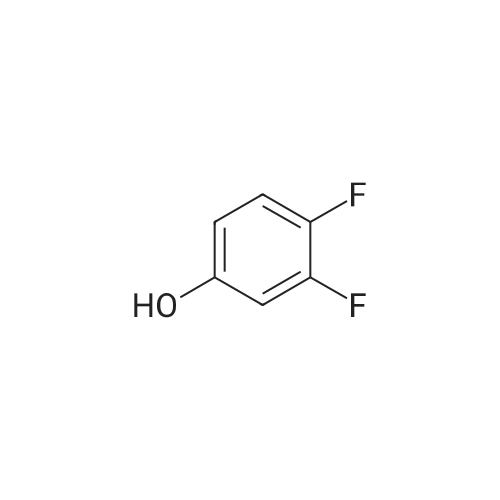
371-41-5 ;
1187431-43-1 ;
60-00-4 ;
3483-12-3
SAR study of piperidine derivatives as inhibitors of 1,4-dihydroxy-2-naph- thoate isoprenyltransferase (MenA) from Mycobacterium tuberculosis
Berg, Kaja
;
Hegde, Pooja
;
Pujari, Venugopal
, et al.
Eur. J. Med. Chem.,2023,249,115125.
DOI:
10.1016/j.ejmech.2023.115125
PubMed ID:
36682292
More
Abstract: The electron transport chain (ETC) in the cell membrane consists of a series of redox complexes that transfer electrons from electron donors to acceptors and couples this electron transfer with the transfer of protons (H+) across a membrane. This process generates proton motive force which is used to produce ATP and a myriad of other functions and is essential for the long-term survival of Mycobacterium tuberculosis (Mtb), the causative organism of tuberculosis (TB), under the hypoxic conditions present within infected granulomas. Menaquinone (MK), an important carrier molecule within the mycobacterial ETC, is synthesized de novo by a cluster of enzymes known as the classic/canonical MK biosynthetic pathway. MenA (1,4-dihydroxy-2-naphthoate prenyltransferase), the antepenultimate enzyme in this pathway, is a verified target for TB therapy. In this study, we explored structure-activity relationships of a previously discovered MenA inhibitor scaffold, seeking to improve potency and drug disposition properties. Focusing our campaign upon three molecular regions, we identified two novel inhibitors with potent activity against MenA and Mtb (IC50 = 13-22 μM, GIC50 = 8-10 μM). These analogs also displayed substantially improved pharmacokinetic parameters and potent synergy with other ETC-targeting agents, achieving nearly complete sterilization of Mtb in combination therapy within two weeks in vivo. These new inhibitors of MK biosynthesis present a promising new strategy to curb the continued spread of TB.
Keywords:
1,4-dihydroxy-2-naphthoate prenyltransferase ;
MenA ;
MenA inhibitors ;
Menaquinone ;
Mtb ;
Mycobacterium tuberculosis ;
Piperidine derivatives ;
SAR
Purchased from AmBeed:
25952-53-8 ;
90719-32-7 ;
872-85-5 ;
6457-49-4 ;
3769-41-3 ;
10338-57-5 ;
135-19-3 ;
135-19-3 ;
28177-48-2 ;
22246-18-0 ;
122334-37-6 ;
91914-06-6 ;
10040-98-9 ;
161975-39-9 ;
150-76-5 ;
371-41-5 ;
63754-96-1 ;
288-32-4 ;
3380-34-5 ;
1677-46-9 ;
166815-96-9 ;
700-57-2 ;
1204-86-0 ;
21725-69-9 ;
367-12-4 ;
1003-29-8 ;
627-35-0 ;
27292-49-5 ;
104324-16-5 ;
123855-51-6 ;
4328-13-6 ;
875401-70-0 ;
405272-71-1 ;
63614-86-8 ;
1420942-13-7 ;
25952-53-8 ;
1420895-21-1 ;
1078-18-8 ;
32363-45-4 ;
69564-68-7 ;
31519-22-9 ;
22246-18-0 ;
189618-33-5 ;
180847-24-9 ;
6264-98-8 ;
946680-75-7 ;
63608-38-8 ;
713-68-8 ;
62810-39-3 ;
189618-32-4 ;
180847-23-8 ;
63608-31-1 ;
15789-05-6 ;
63712-27-6 ;
63608-33-3 ;
63608-35-5
...More

 Chemistry
Chemistry
 Pharmaceutical Intermediates
Pharmaceutical Intermediates
 Inhibitors/Agonists
Inhibitors/Agonists
 Material Science
Material Science














 For Research Only
For Research Only
 120K+ Compounds
120K+ Compounds
 Competitive Price
Competitive Price
 1-2 Day Shipping
1-2 Day Shipping