|
|
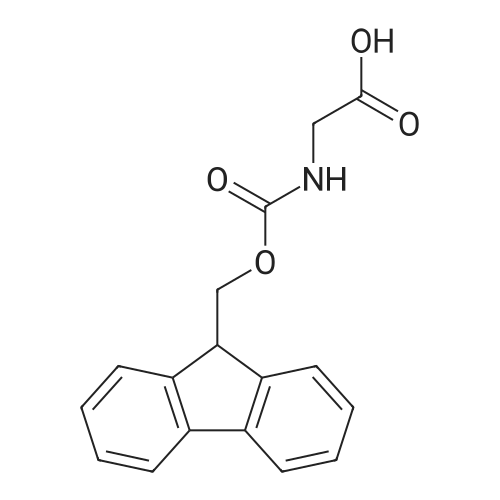
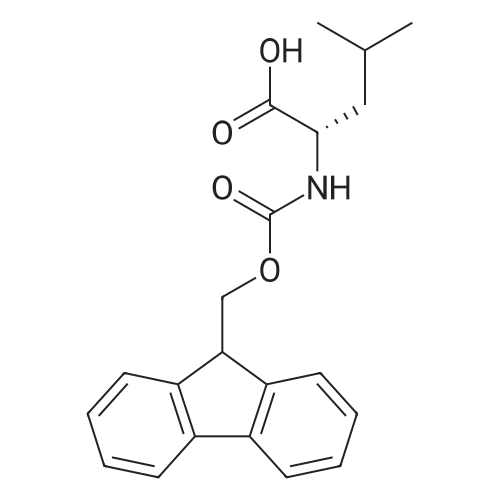
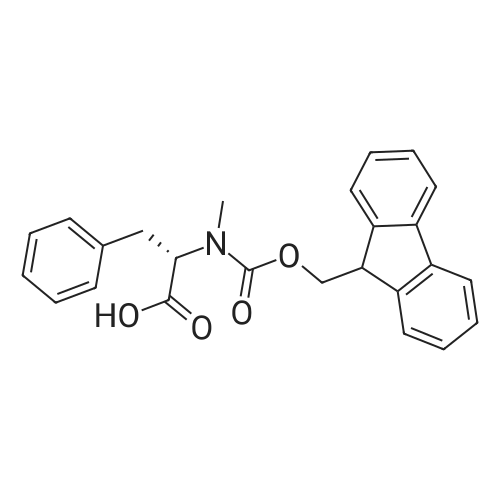
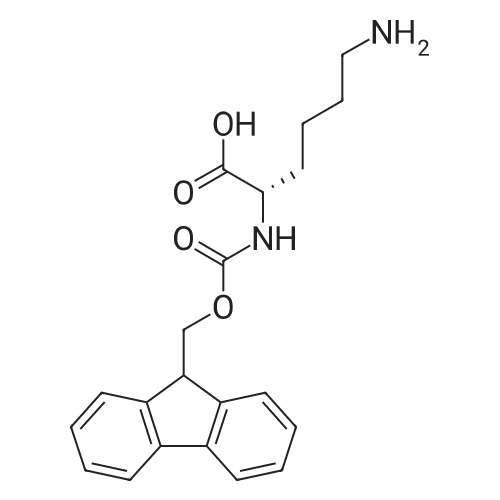
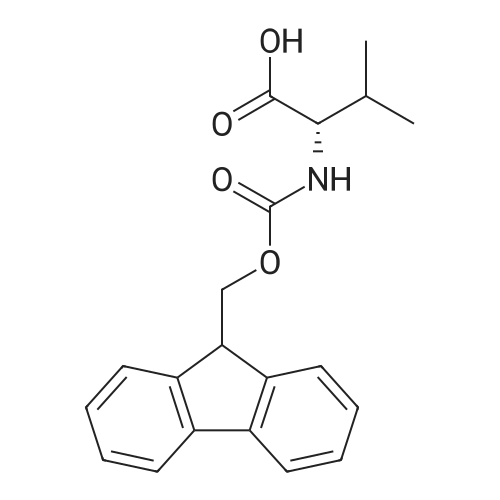
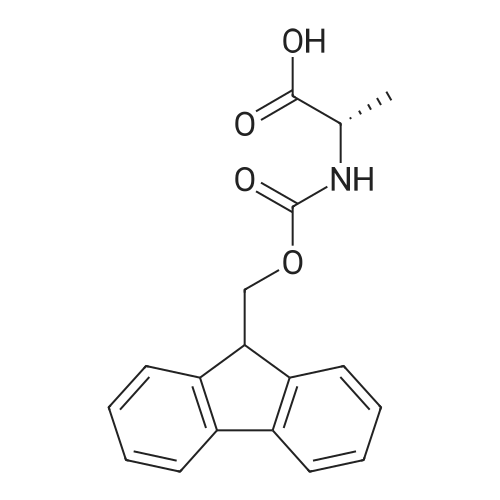
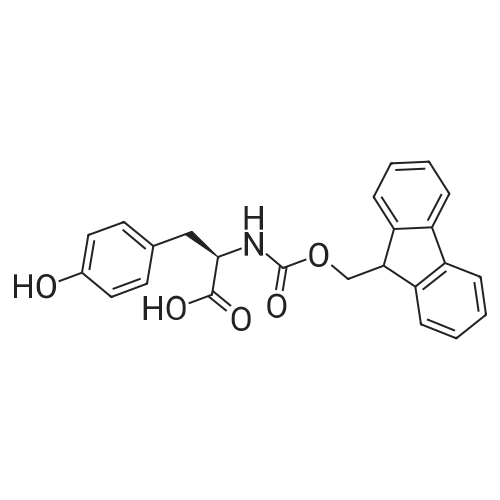
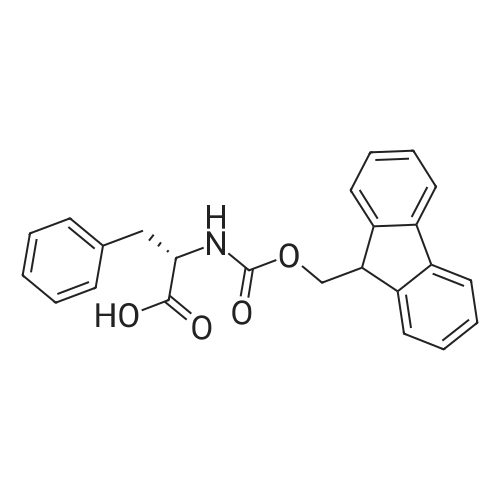
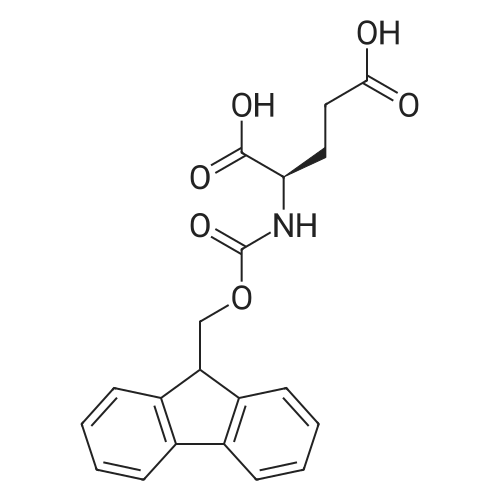
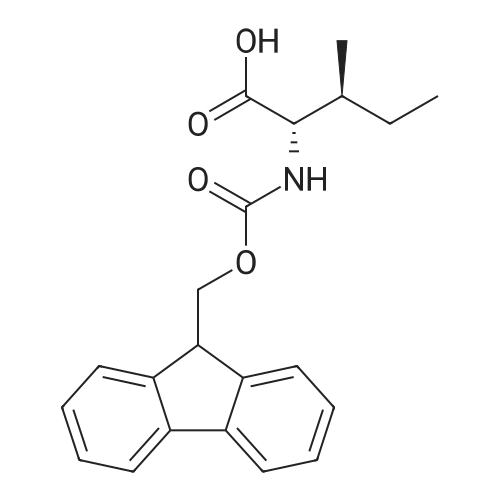
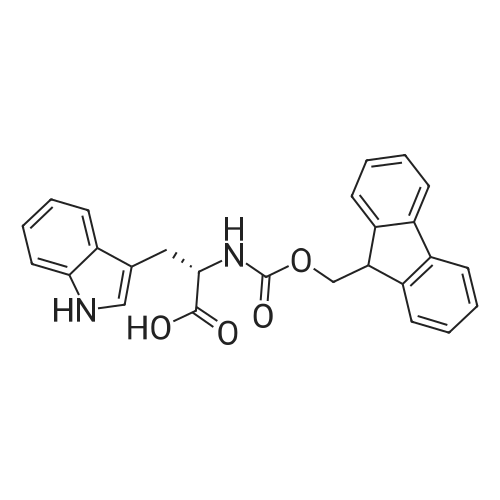
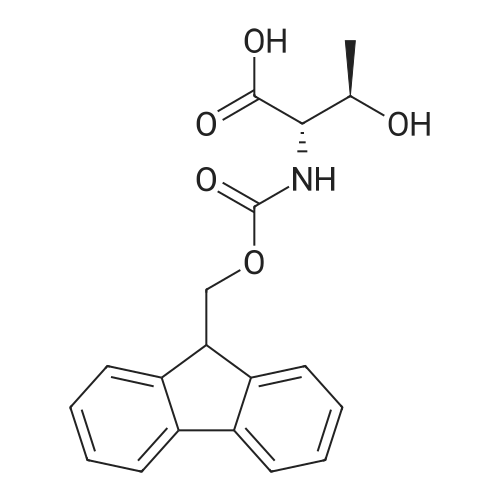
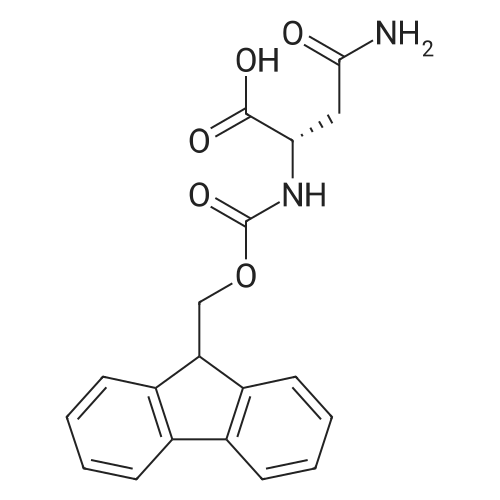
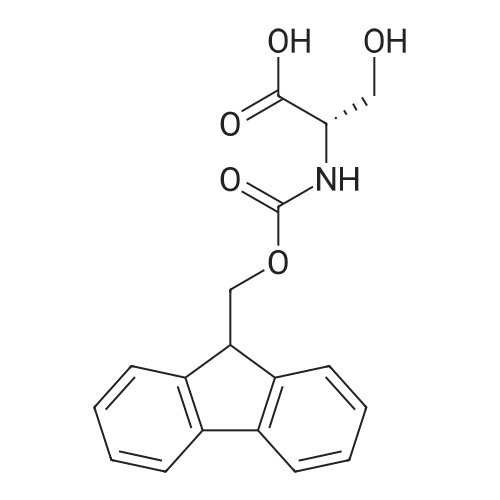
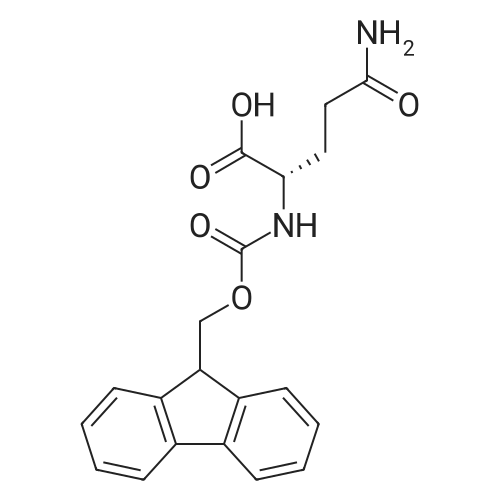
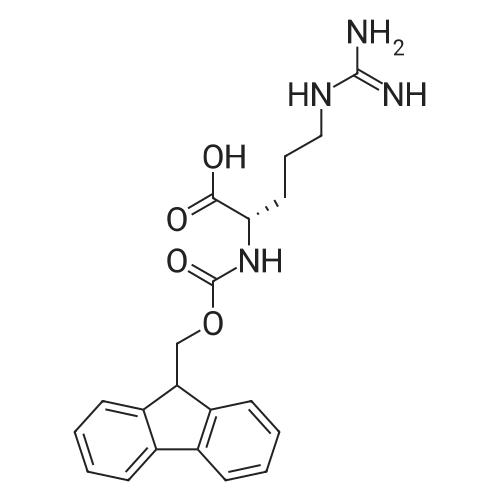
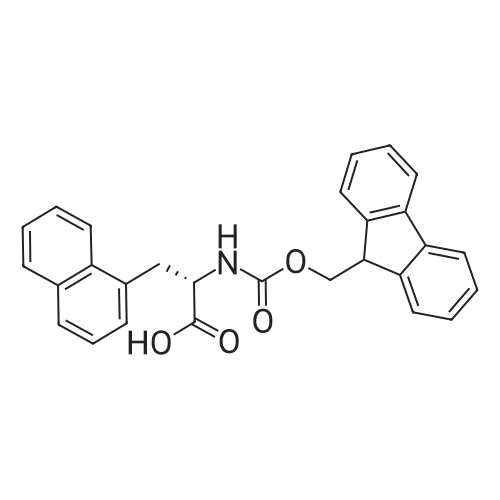
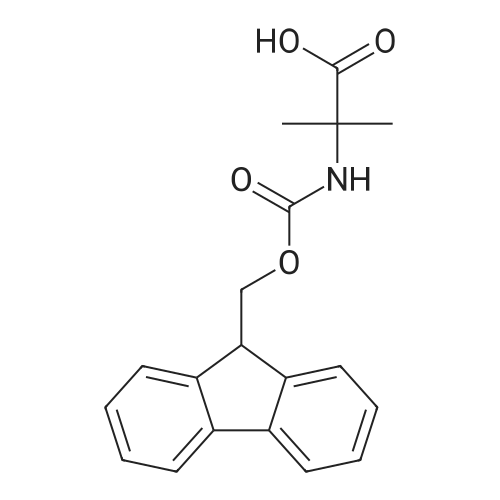
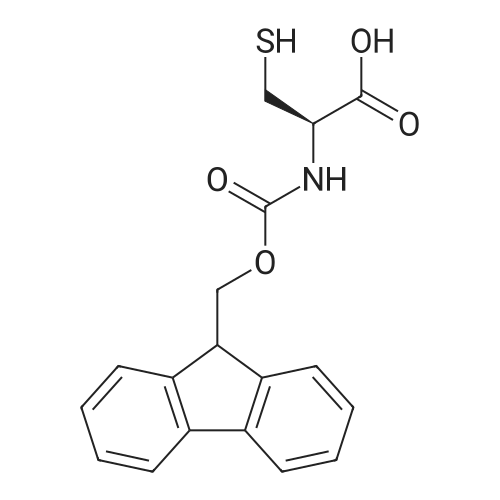
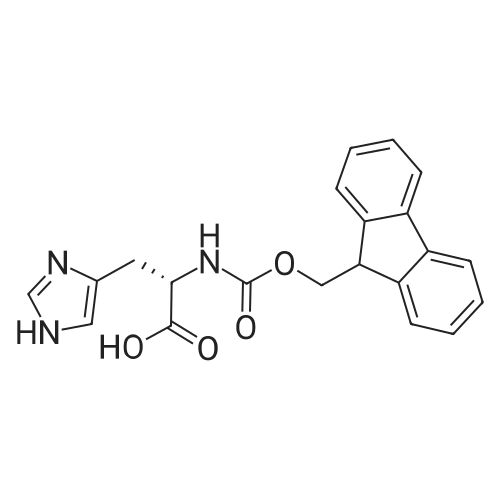
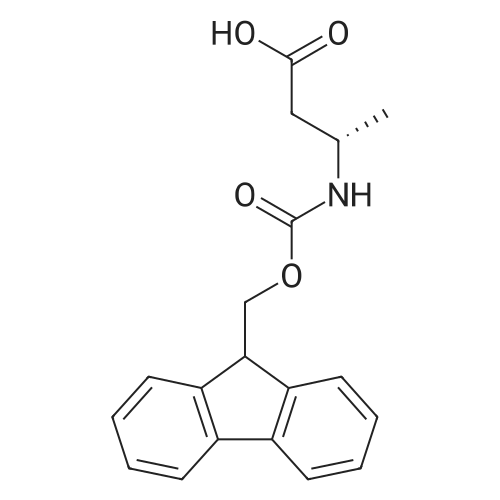
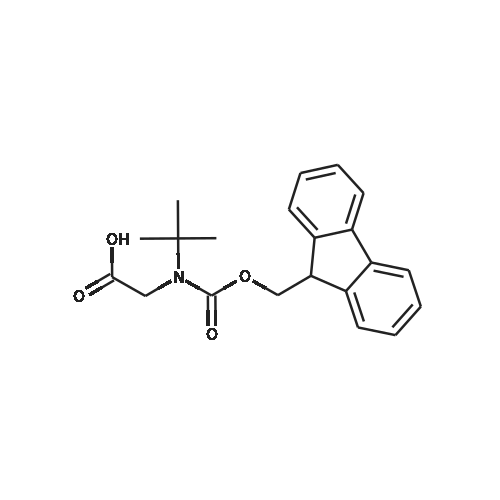
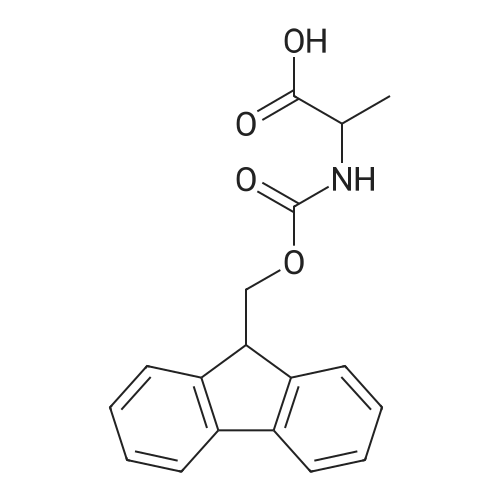
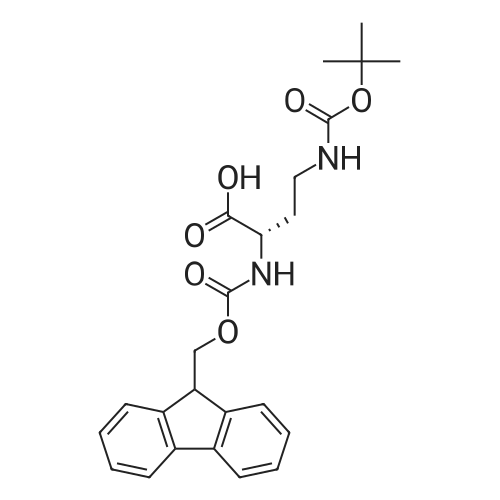
1. Peptide synthesis 1.1 General synthetic procedures A general method for the synthesis of the peptidomimetics of the present invention is exemplified in the following. This is to demonstrate the principal concept and does not limit or restrict the present invention in any way. A person skilled in the art is easily able to modify these procedures, especially, but not limited to, choosing a different starting position within the ring system, to still achieve the preparation of the claimed cyclic peptidomimetic compounds of the present invention. Coupling of the first protected amino acid residue to the resin . In a dried flask, 2-chlorotritylchloride resin (polystyrene, 1percent crosslinked; loading: 1.4 mMol/g) was swollen in dry CH2CI2 for 30 min (7 mL CH2CI2 per g resin). A solution of 0.8 eq of the Fmoc-protected amino acid and 6 eq of DIPEA in dry CH2CI2/DMF (4/1) (10 mL per g resin) was added. After shaking for 2-4 h at rt the resin was filtered off and washed successively with CH2CI2, DMF, CH2CI2, DMF and CH2CI2. Then a solution of dry CH2CI2/MeOH/DIPEA (17:2:1) was added (10 mL per g resin). After shaking for 3 x 30 min the resin was filtered off in a pre-weighed sinter funnel and washed successively with CH2CI2, DMF, CH2CI2, MeOH, CH2CI2, MeOH, CH2CI2 (2x) and Et20 (2x). The resin was dried under high vacuum overnight. The final mass of resin was calculated before the qualitative control. Loading was typically 0.6 - 0.7 mMol/g. The following preloaded resins were prepared: Fmoc-Dab(Boc)-2-chlorotrityl resin, Fmoc-DDab(Boc)-2-chlorotrityl resin, Fmoc-Lys(Boc)-2-chlorotrityl resin, Fmoc- Trp(Boc)-2-chlortrityl resin, Fmoc-Phe-2-chlortrityl resin; Fmoc-Val-2-chlorotrityl resin, Fmoc-Pro-2-chlorotrityl resin, Fmoc-Arg(Pbf)-2-chlorotrityl resin and Fmoc-Glu(iBu)-2- chlorotrityl resin. Synthesis of the fully protected peptide fragment The synthesis was carried out on a Syro-peptide synthesizer (MultiSynTech GmbH) using 24 to 96 reaction vessels. In each vessel 0.04 mMol of the above resin were placed and the resin was swelled in CH2CI2 and DMF for 15 min, respectively. The following reaction cycles were programmed and carried out: Step Reagent Time 1 CH2CI2, wash and swell (manual) 1 x 3 min 2 DMF, wash and swell 2 x 30 min 3 20percent piperidine/DMF 1 x 5 min and 1 x 15 min 4 DMF, wash 5 x 1 min 5 3.5 eq Fmoc amino acid/3.5 eq HOAt in DMF + 3.5 eq PyBOP/7 eq DIPEA or 3.5 eq DIC 1 x 40 min 6 3.5 eq Fmoc amino acid/DMF + 3.5 eq HATU or PyBOP or HCTU + 7 eq DIPEA 1 x 40 min 7 DMF, wash 5 x 1 min 8 20percent piperidine/DMF 1 x 5 min and 1 x 15 min 9 DMF, wash 5 x 1 min 10 CH2CI2, wash (at the end of the synthesis) 3 x 1 min Steps 5 to 9 are repeated to add each amino-acid residue. After the termination of the synthesis of the fully protected peptide fragment, one of the procedures A - E, as described herein below, was adopted subsequently, depending on which kind of interstrand linkages, as described herein below, were to be formed. Finally, the peptides were purified by preparative reverse phase LC-MS, as described herein below. Procedure A: Cyclization and work up of a backbone cyclized peptide having no interstrand linkage Cleavage, backbone cyclization and deprotection After assembly of the linear peptide, the resin was suspended in 1 mL of 1percent TFA in CH2CI2 (v/v; 0.14 mMol) for 3 minutes. After filtration the filtrate was neutralized with 1 mL of 20percent DI PEA in CH2CI2 (v/v; 1.15 mMol). This procedure was repeated four times to ensure completion of the cleavage. An alternative cleavage method comprises suspension of the resin in lmL of 20percent HFIP in CH2CI2 (v/v; 1.9 mMol) for 30 minutes, filtration and repetition of the procedure. The resin was washed three times with 1 mL of CH2CI2. The CH2CI2 layers containing product were evaporated to dryness. The fully protected linear peptide was solubilised in 8 mL of dry DM F. Then 2 eq of HATU and 2 eq of HOAt in dry DM F (1-2 mL) and 4 eq of DIPEA in dry DM F (1-2 mL) were added to the peptide, followed by stirring for ca. 16 h. The volatiles were removed by evaporation. The crude cyclic peptide was dissolved in 7 mL of CH2CI2 and washed three times with 4.5 mL 10percent acetonitrile in water (v/v). The CH2CI2 layer was then evaporated to dryness. To fully deprotect the peptide, 7 mL of cleavage cocktail TFA/DODT/thioanisol/H20 (87.5 :2.5:5:5) or TFA/TIS/H20 (95:2.5 :2.5) were added, and the mixture was kept for 2.5-4 h at room temperature until the reaction was completed. The reaction mixture was evaporated close to dryness, the peptide precipitated with 7 mL of cold Et20/pentane and finally washed 3 times with 4 mL of cold Et20/pentane. Procedures Bl and B2: Cyclization and work up of a backbone cyclized peptide having a disulfide interstrand linkage Bl: Formation of a disulfide interstrand linkage using DMSO After cleavage, backbone cyclization and deprotection of the linear peptide, as described in the corresponding section of procedure A, th... |

 Chemistry
Chemistry
 Pharmaceutical Intermediates
Pharmaceutical Intermediates
 Inhibitors/Agonists
Inhibitors/Agonists
 Material Science
Material Science














 For Research Only
For Research Only
 120K+ Compounds
120K+ Compounds
 Competitive Price
Competitive Price
 1-2 Day Shipping
1-2 Day Shipping